MULTIOMICS CORE
MULTIOMICS CORE
The core functions are to promote multiomics approaches to skin biology and disease research, enabling investigators to dissect molecular and cellular heterogeneities within skin to predict lineages, cell state transitions, and cell-cell interactions. Specifically, the core supports genomic DNA sequencing technologies: bulk, single cell, and spatial transcriptomics, as well as bioinformatic analysis of sequencing data across these platforms.
Core Leadership
Suzanne Sandmeyer, PhD
Director
https://genomics.uci.edu/
sbsandme@uci.edu
Maksim Plikus, PhD
Associate Director
https://plikuslab.bio.uci.edu/
plikus@uci.edu
Melanie Oakes, PhD
Manager
mloakes@uci.edu
Jenny Wu, PhD
Director of Bioinformatics
jiew5@uci.edu
Ivan Chang, PhD
Research Computing
iychang@uci.edu
Cindy Yamamoto, PhD
Research Specialist
cyamamo1@hs.uci.edu
The core is directed by Suzanne Sandmeyer, PhD, an expert in genomics technologies and co-directed by Max Plikus, PhD, an expert in skin molecular biology. Jenny Wu, PhD is Director for Bioinformatics and specializes in multiple aspects of omics analysis. Ivan Chang, PhD, specializes in bioinformatic engineering and design of data sharing systems and interfacing of the core with the high-performance computing cluster (HPC3).
Contacts: Users, particularly new users, are encouraged to make an appointment with Oakes (mloakes@uci.edu) and Wu (jiew5@uci.edu) in advance of project execution in order to discuss experimental strategy and statistical validation, respectively, of anticipated data. Oakes may be reached at (949) 824-6023 and her office is Sprague 336. Lab staff may be reached directly at (949) 824-5327. Additional information regarding submitting orders is available on the Genomics Research and Technology Hub Genomics Research and Technology Hub Website and at the iLab link.
MULTIOMICS RESEARCH SUPPORT
Library construction - The staff has experience in Illumina Nova Seq X Plus short read sequencing and PacBio Revio long read sequencing for bulk or single cell RNA analysis. Depending on client needs, the facility can prepare manually or using automation, mRNA, small RNA, genomic and exome sequencing libraries. The staff has experience with different strategies for exome enrichment, ribosomal RNA depletion and generation of multiplex libraries to insure maximal return of data in your experiments. In addition to internal library processing, externally prepared libraries are subjected to quality control measurements prior to sequence analysis. To facilitate the preparation of libraries and quality testing and final titration of samples the GRT Hub is equipped with Covaris S220 focused ultrasonicator; NanoDrop 1000 and NanoDrop One spectrophotometers, Bio-Rad CFX 96 thermal cyclers, Agilent 2100 Bioanalyzer, Sage Biosciences Blue Pippin and ThermoFisher Quantstudio 7 flex and Agilent Femto Pulse. A Hamilton STAR instrument has been recently acquired to automate preparation of frequently used libraries.
Gene expression and single cell analysis - The core offers services that target applications for single cell genomics using the 10x Genomics Chromium, Parse Biosciences Combinatorial Barcoding, Illumina Pip-Seq, Mission Bio Tapestri, and Bruker IsoSpark platforms. Single cell genomic approaches including 3’ gene expression, immune profiling, ATAC-seq, sample multiplexing, protein detection, and multiomics (e.g., RNA- with protein or ATAC-seq) workflows are available to support skin research.
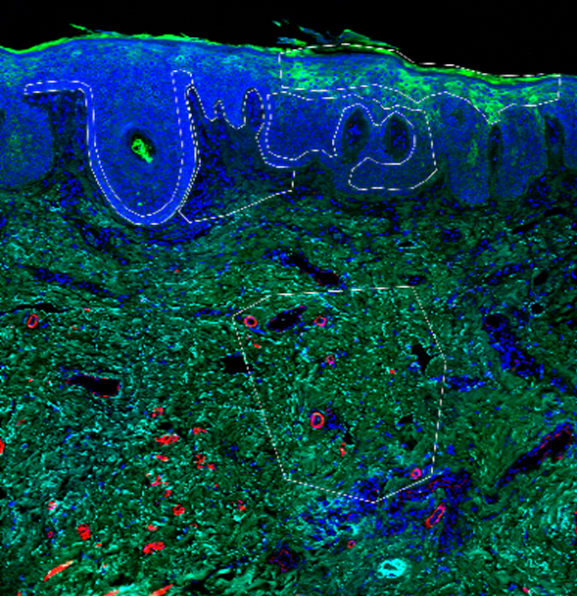
Spatial transcriptomics - The core supports spatial analysis on 10X Genomics CytAssist enabled Visium HD for discovery and Xenium for subcellular probe-based approaches and NanoString GeoMx protein and RNA probing, as well as RNAScope for validation studies. Platforms accommodate both fresh frozen and FFPE tissues, with the latter enabling spatial transcriptomics on clinically archived skin samples. See image from Kraus and Dai above with clinical application for lichen sclerosis samples.
Genomics Technologies
The Multiomics Core instrumentation includes the following technologies:
Illumina NovaSeq X Plus - Genome assembly, structural variation, variant analysis, differential gene expression, SNP discovery, DNA-protein interaction (ChIP-Seq), methylation analysis, small RNA discovery and analysis.
Illumina MiSeq i100 - The Illumina MiSeq system has a wide variety of focused applications: targeted gene sequencing, small genome sequencing, 16S metagenomics, amplicon sequencing, etc. The unique chemistry of the MiSeq allows for sequencing up to 2×300 bp read length and 2 x 500 bp on the MiSeq i100+.
PacBio Revio - The PacBio Revio is based on a novel single-molecule, real-time technology which enable the observation of natural DNA synthesis by a DNA polymerase in real time. The sequencing occurs on SMRT cells each containing 8M zero-mode waveguides (ZMWs). The ZMWs provide a window for watching the DNA polymerase as it performs sequencing by synthesis. The Revio is amplification free and capable of multi-kilobase reads. Applications include de novo assembly, analysis of epigenetic modifications and characterization of genomic variation including repeat expansions, isoform analysis, compound mutations and haplotype phasing and SNP detection and validation. The Revio enables MAS-Seq determination of single cell iso-form RNAs.
NanoString nCounter - NanoString’s Prep Station and MAX analysis system uses color-coded molecular barcodes that can hybridize to directly to different types of target molecules. This digital detection technology is capable of direct profiling of individual molecules in a single reaction without amplification. RNA analysis applications include gene expression, single cell gene expression, lncRNA expression, miRNA expression and Leukemia fusion gene analysis.
10X Genomics Chromium X and CytAssist - The Chromium controller can encapsulate individual cells or genomic DNA into addressable partitions, each with an identifying barcode for downstream analysis. The proprietary GemCode technology utilizes a droplet-based system to capture from 100-100,000 cells in a 10-minute run. Up to 8 samples can be processed in parallel. Chromium X is a flexible instrument and can run all single cell assays, including new high-throughput assays for Single Cell Immune Profiling and Single Cell Gene Expression.
NanoString GeoMx - NanoString’s GeoMx Digital Spatial Profiler (DSP) combines the best of spatial and molecular profiling technologies by generating digital whole transcriptomes and profiling data for 100s of validated Protein analytes from up to 12 tissue slides per day. This unique combination of high-plex and high-throughput spatial profiling enables researchers to rapidly and quantitatively assess the biological implications of the heterogeneity within tissue samples.
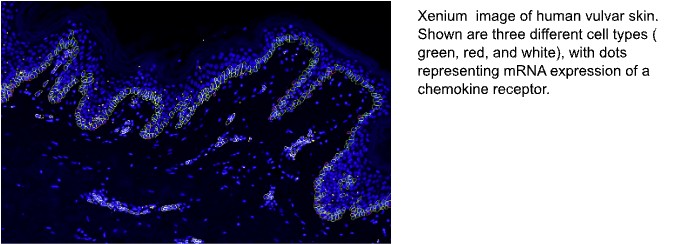
10X Genomics Xenium enables visualization of up to 500 different types of transcripts at subcellular resolution. It can be used with FF or FFPE tissue. Hybridization and imaging is automated. Visualization is based on a padlock probe system in which only split probes able to be ligated based on juxtaposition on target transcripts are subject to rolling circle amplification. Fluorescently coded probes are then used to read out the transcripts. Multiple probes are used per gene. 10x Genomics segmentation is currently DAPI based, however open source methods are becoming more sophisticated. Proteins can also be tagged and localized in the same sample. 10x Genomics recently launched the Xenium Cell Segmentation package that provides researchers with improvements in assignment of transcripts to cells. The data is delivered immediately when the run is complete, and is a result of optimized membrane, interior, and nuclear stains combined with a machine learning algorithm that optimizes use of the available signal across multiple channels.
Vizgen Merscope Ultra introduces combinatorial barcoding at the subcellular level with FOV up to 3cm^ 2 and probe sets up to 1000. Now available for both the MERSCOPE Ultra™ and the MERSCOPE® Platform, MERFISH 2.0 leverages enhanced RNA anchoring and signal enhancement to build on the original MERFISH sensitivity and reproducibility
Support instruments for sample extraction, QC, library preparation and genotyping include - Illumina iScan bead arrays for SNP, CNV, and methylation analysis, Apollo system for library preparation, Formulatrix Mantis liquid dispenser, KingFisher DNA extraction system, Covaris S220 focused Acoustic Shearer, Agilent Bioanalyzer 2100 and Mx3005 Real-Time PCR instrument, QuantStudio 7 Flex, real-time PCR, NanoDrop ND-1000 and NanoDrop One spectrophotometers, and Speed Vac concentrator system.
Bioinformatics
Analysis tools - In addition to specialized analysis with state-of-the-art open-source tools, several commercial software programs including Genomics Workbench (CLC Bio) and JMP Genomics (SAS) are available to P30 skin researchers via licensing. Jenny Wu, Ph.D., Director of Bioinformatics service at the core, is available on a recharge basis to assist users with data analysis for sequencing, microarray, and spatial omics projects. Dr. Wu has extensive experience with approaches for sc/snRNA-seq as well as bulk RNA analysis. In particular for spatial analysis, the core continues to support state-of-the-art single-cell analytic tools and has started adding new spatial omics tools on the internal data analysis portal including Tangram, CellChat, COMMOT, Banksy, spacexr (RCTD), cellpose and Giotto etc., available to all center researchers. Computational tools such as Tangram) and CellChat aim to combine high throughput molecular data (e.g., scRNA-seq) with spatially resolved omics data (e.g., Visium, Merscope, etc.) to derive biological insights. Interactive analysis and visualization tools such as spatialLIBD and Giotto provide user friendly functionalities and allow for visualization of continuous and categorical measurements along with the tissue images. The core collaborates with investigators for example using CellChat and other tools developed by the Nie laboratory in the Systems Core.
Data sharing - Bioinformatics Director Wu and Bioinformatics Engineer Chang actively support users with open-source and commercial tools available in the core’s SkinGenes portal via the UCI HPC3 dedicated node. Standard open-source tools include Seurat, Monocle, Giotto, DESeq2, limma, Bowtie2, Samtools, IGV etc. On the visual computing interface front, the tailored Jupyterlab scientific computing framework of the internal hub will provide P30 skin researchers: shared interactive Python or R Jupyter notebooks documenting specific analysis methods and results and curated bioinformatics analysis pipelines and workflow packaged in reproducible singularity containers; Available tools on the web portal include CellChat, COMMOT developed by Plikus and Nie groups. Tangram and spacexr tools have been containerized and hosted on the data portal for easy access, improved re-usability and reproducibility of the workflow. SpatialLIBD is hosted on a web-based R shiny server for interactive spatial omics data sharing and exploration, available to all center researchers. Contact Ivan Chang, PhD, iychang@uci.edu for inquiries regarding posting new data sets to the portal or for technical issues with the portal.

Bulk RNA sequencing workshop,
Jenny Wu, PhD, Ivan Chang, PhD,
and Fabio Macciardi, PhD, UCI
Workshops, Tech Talks, and Seminars
The core cooperates with UCI Genomics Research and Technology Hub (GRT Hub), managed by Oakes, in hosting vendor technical seminars, and updates on technology available in the GRT Hub. Additionally, Wu and Chang conduct a series of in-person bioinformatics workshops in alternate months.
This year those workshops have included introduction to the HPC3 and Linux, bulk RNA sequencing analysis of gene expression, single cell RNA-seq and statistics and deep learning in genomics.
Future workshops and recordings of many past workshops, tech talks, and seminars can be found on the GRT Hub website:
https://www.genomics.uci.edu/.
-
Hands-On Data Analysis workshop on June 27, 2023. This is mainly one-on-one instruction.
-
DNA Methylation Data Analysis workshop on April 25, 2023. The links to the presentations are as follows:
-
Introduction to DNA Methylation Data Analysis, Jenny Wu, PhD - (PDF)
-
Using Oxford Nanopore for Long Read Sequencing, Jaz Sakr, GSR - (PDF)
-
Introduction to DNA Methylation Data Analysis, Jenny Wu, PhD - Presentations (Videos)
-
Using Oxford Nanopore for Long Read Sequencing, Jaz Sakr, GSR - Presentations (Videos)
-
-
Single Cell RNA-seq Data Analysis Workshop on January 31, 2022, and February 7, 2022. The links for the presentations are as follows:
-
Hands-on Workshop for bulk RNA-seq on September 7, 2022 (ISEB, Room 1010). The links for the presentations are as follows:
-
Bioinformatics Workshop on June 27, 2022 - Introduction to Linux, HPC3 and R – The links for the presentations are as follows:
Also available are the links to the step-by-step guides for the workshop materials via the following links.
- Intro to Bioinformatics Analysis:
https://docs.google.com/document/d/1HQUCqPMMjwUT2fhlC9jyYgqOrjwVKiLJLpR4BjK6yKc/edit?usp=sharing
- Bulk RNA-seq Pipeline:
https://docs.google.com/document/d/1YtKsTTQuS0bYSJBiQ-jbDqcg-E8PnIhM_YEFwO0I2Sw/edit?usp=sharing





